Tagged: 5.13

Recombinant DNA: Grade 9 Understanding for IGCSE Biology 5.12 5.13 5.14
In the last post on this topic, I explained about the two types of enzymes needed for the genetic modification of organisms:
Restriction enzymes that can cut up DNA molecules at specific target sequences, often resulting in fragments with sticky ends
DNA ligase that joins together fragments to form a single DNA molecule
The EdExcel iGCSE syllabus uses the example of the genetic modification of bacteria to produce human insulin. Human insulin is a hormone that helps regulate the concentration of glucose in the blood. It is made in the pancreas when the blood glucose concentration gets too high and causes liver cells to take up glucose from the blood and convert it to the storage molecule glycogen. Patients with type I diabetes cannot make their own insulin and so need to inject it several times a day after meals to ensure they maintain a constant healthy concentration of glucose in their blood.
Bacteria can be genetically modified so that they produce human insulin. These transgenic bacteria can be cultured in a fermenter and the insulin produced can be extracted, purified and sold.
How do you get hold of the human insulin gene?
Well the honest answer is that there are a variety of ways of achieving this. It can now be synthesised artificially as we know the exact base sequence of the gene but it can also be cut out of a human DNA library using a restriction enzyme. There are other ways too but for the sake of brevity (and sanity) I am not going to go into them here. [If you are really interested in this, find out how reverse transcription of messenger RNA from cells in the pancreas can allow you to build the insulin gene.]
How do you get the human insulin gene into a bacterium?
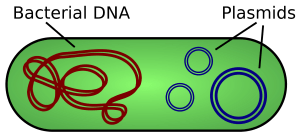
Remember that bacterial cells are fundamentally different to animal and plant cells. One difference is that bacterial cells have no nucleus and their DNA is in the form of a ring that floats in the cytoplasm. Many bacteria also have plasmids which are small additional rings of DNA and these provide a way for getting a new gene into a bacterium.
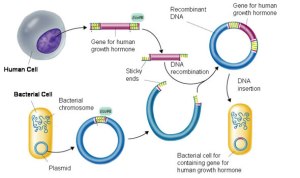
Bacteria exchange plasmids in a process called conjugation and so it is fairly easy to get the plasmid out of the bacterium. If the plasmid is cut open using the same restriction enzyme as was used to cut out the human insulin gene, the sticky ends will match up and so DNA ligase will join the two pieces of DNA together to make a recombinant plasmid. The diagram below shows the process for human growth hormone but it would be exactly the same for the example we are looking at.
If the recombinant plasmids are inserted into bacteria, the bacteria will read the human insulin gene and so produce the protein insulin.
How do you grow the transgenic bacteria on an industrial scale?
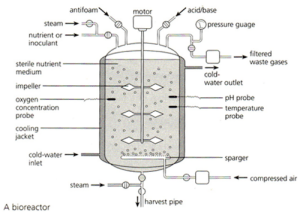
The bacteria that have taken up the recombinant plasmid are grown in a fermenter. This is a large stainless steel vat (easy to clean and sterilise) that often has several design features conserved between different varieties:
The fermenter usually has a cooling jacket to carry away excess heat. The jacket often has a cold water input pipe and the warmer water is carried away. There has to be some mechanism for mixing the contents of the fermenter so the diagram above shows paddles attached to a motor. Fermenters also need a sterile input system for getting air, water and nutrients into the fermenter but without introducing foreign bacteria and fungi. Air is needed as the bacteria are aerobic and need oxygen for respiration.
If the bacteria in the fermenter contain the human insulin gene, then they will be able to produce human insulin. This can be extracted, purified and sold to the NHS for treating type I diabetics.

Genetic Modification Grade 9 Understanding for IGCSE Biology 5.12 5.13 5.14 5.16
One of the most complicated areas in the iGCSE course is looking at how organisms can be genetically modified. Remember that humans have been messing around with the genetic composition of many species for thousands of years. Up until recently this has only been using a technique called selective breeding or artificial selection.
Make sure you understand exactly what is meant by the term selective breeding? You probably should be able to explain at least one example in both an animal and a plant species.
In the twentieth century scientists developed a much more precise way of genetically modifying a species. This was due to discoveries about the nature of the genetic code and also the existence of two types of enzyme that make cutting up and then sticking together pieces of DNA. This new technique was called genetic engineering and it has two big advantages over selective breeding. Firstly genes from different species can be recombined to form transgenic organisms. Transgenic is an important term and means an organism that contains DNA from more than one species. This means scientists are not restricted to alleles present in the natural population but can insert genes from any species into any other. Secondly, selective breeding has a big disadvantage in that it can reduce the allelic diversity in a population. If the population becomes more and more similar at a genetic level, this means that inbreeding becomes more of a problem and the population becomes susceptible to damage from changes in the environment.
You need to understand the role of two enzymes in the process of Genetic Engineering:
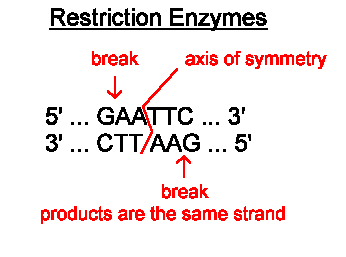
Restriction Enzymes are found in bacteria and have evolved to combat viral infection in the bacterial cell. These enzymes can cut double-stranded DNA at a specific target sequence, often leaving the ends of the DNA with short sections of unpaired bases. These are called sticky ends.
The image below shows the cutting site of a restriction enzyme called EcoR1. You can see the enzyme cuts the DNA anywhere the following sequence is found GAATTC. The DNA molecule is cut after the first G, leaving the two strands with four unpaired bases that make up the two sticky ends.
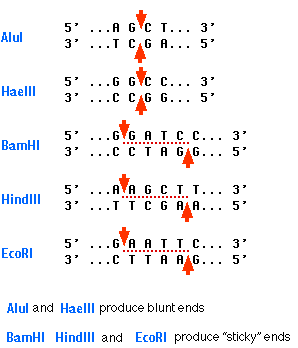
Here are some more restriction enzymes and there target sequences. The bottom three on the list all produce sticky ends.
The second important enzyme for genetic engineering is DNA Ligase. This enzyme catalyses the joining together of the sticky ends of two fragments to produce an intact DNA molecule.
The starting point for understanding the complex topic of genetic modification of organisms is understanding the role of these two enzymes in the process.