Tagged: anticodon
Protein Synthesis (part 3): Grade 9 Understanding for IGCSE Biology 3.18B
This is my final post on protein synthesis, you may be relieved to know…. It is a complicated topic and there is lots to understand but remember it is only one specification point out of several hundred in the IGCSE specification…… So don’t worry too much if you find this hard to grasp and don’t spend a disproportionate amount of time revising it. If you are fascinated by how genetic information is encoded in DNA and how genes work at a molecular level, then you have to choose Biology as a subject to study at A level!
I want to end with two final questions, both of which are essential to address if you are to acquire the grade 9 understanding you need.
What is meant by complementary base pairing and why is it so important?
Let’s go back to the structure of a DNA molecule.
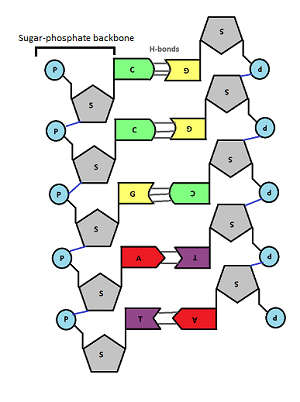
In the middle of the molecule there are pairs of bases. There are four possible bases in DNA – Adenine, Thymine, Cytosine and Guanine – but they are often represented by their letters A,T,C and G.
Bases pair up in a totally predictable way across the double stranded DNA molecule:
A always pairs with T, C always pairs with G. Why is this? Well you can see from the diagram above that A and T are held together by two weak bonds called Hydrogen (H) bonds, whereas C and G are held together by 3 H bonds. This means that this is the only way they can pair up in a stable way.
These pairs of bases (A=T and C≡G) are called complementary base pairs because they always match up in a predictable way.
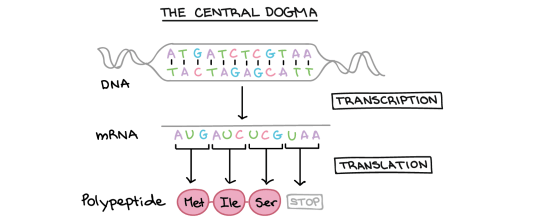
You can also get complementary base pairing between RNA bases. (Remember that in RNA, there is never any thymine but it is replaced with a different base called uracil) So A=U and C≡G are the complementary base pairs between two RNA molecules.
Complementary base pairing between DNA and RNA bases is essential in transcription but you do not need to know the details at this stage.
We will come back to complementary base pairing in a moment…..
How does the ribosome ensure that the correct amino acids are joined together to make the protein during translation?
To understand this, you need to understand the role that transfer RNA (or tRNA for short) plays in protein synthesis. tRNA molecules are found in the cytoplasm and have an interesting structure. At one end, they have an important triplet of bases called the anticodon. At the other end, there is a place in the molecule where an amino acid can be added.

The key idea is this: the tRNA molecules are loaded up by attaching an amino acid by a group of enzymes found in the cytoplasm. But these enzymes ensure that each amino acid is only attached to tRNA molecules with the correct anticodon.
Transfer RNA molecules are so called because they are going to transfer (or carry) the amino acids into the ribosome. You do not need to know the details of protein synthesis but the main idea is that there is complementary base pairing between the anticodon on the tRNA and the codon on the mRNA.
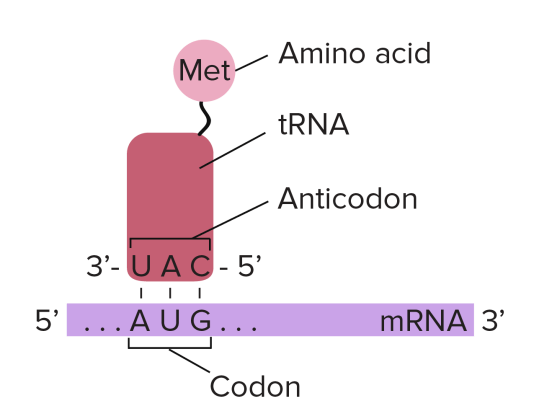
This complementary base pairing between codon and anticodon when combined with the structure of the ribosome means that the amino acids will be joined together in the correct order to make the protein.
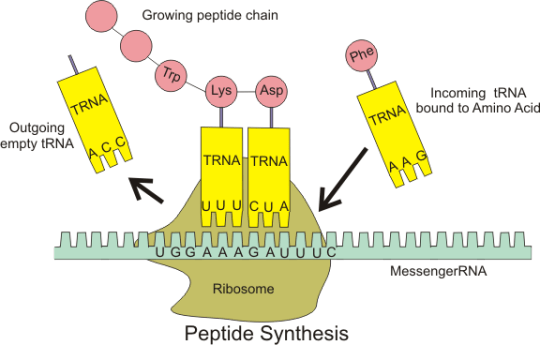
Please do not worry too much about the details of all this. I am sure that the examiners will not expect you to know the details of protein synthesis. But you should understand what an an anticodon is and the role of tRNA in the overall process.
anticodon: “a triplet of adjacent bases found on a transfer RNA molecule”
tRNA: “these molecules are found in the cytoplasm of cells and carry the correct amino acid into the ribosome. There is complementary base pairing between the codon on the mRNA and the anticodon on the tRNA as shown in the diagram above”